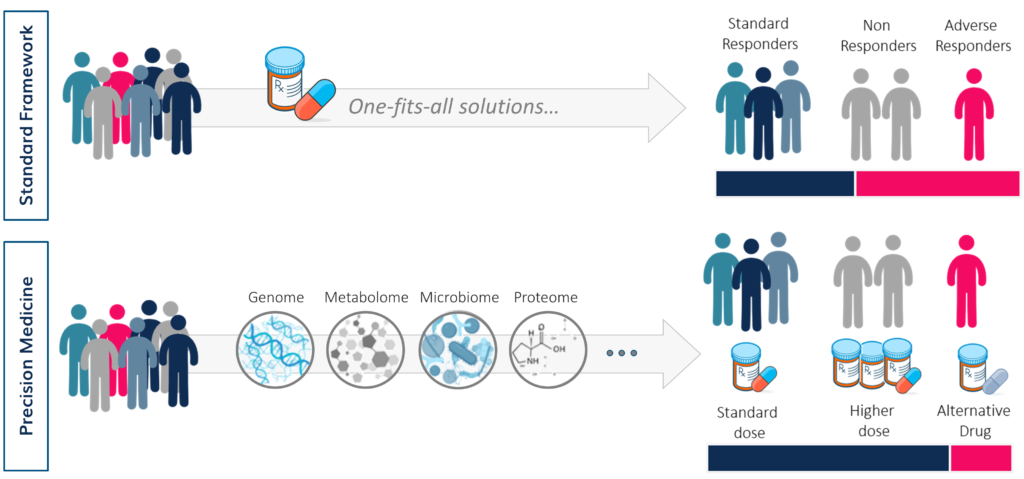
Precision medicine is an emerging approach for disease treatment and prevention that takes into account individual variability in genes, environment and lifestyle. This novel framework is opposed to the traditional practice that develops one-fits-all treatments for populations and make the same medical decisions based on few similar physical and clinical characteristics among patients.
Indeed, precision medicine often involves the analysis of pan-omic (genomic, transcriptomic, metabolomic, gut microbiome, etc.) data to identify the cause of single patients’ disease at the molecular level, classifying individuals into subpopulations that differ in their susceptibility to a particular disease, in the biology or prognosis of the diseases they may develop, or in their response to a specific treatment. Preventive or therapeutic interventions can then be concentrated on those who will benefit, sparing expense and side effects for those who will not.
In this shift and cultural transformation, Machine Learning (ML) and Artificial Intelligence (AI) are the key methodological frameworks that can bring this opportunity to everyday practice.
Indeed, these rich sources of biological information carry complexities such as hyper-dimensionality on small samples, spatial and functional correlation, sparsity, low signal-to-noise ratio and so on, that hinder the applicability of most traditional biostatistical models and methods based on assumptions that are now failing. Conversely, Machine Learning, Deep Learning, Graph and Complex Networks theory, once appropriately tailored to this peculiar setting via careful mathematical choices and biological expertise, can represent powerful tools to extract valuable insights from this rich yet challenging data.
Research projects / lines:
- EPIC1 – Deep Non-Linear EWAS for Breast Cancer Time-to-Diagnosis Modeling
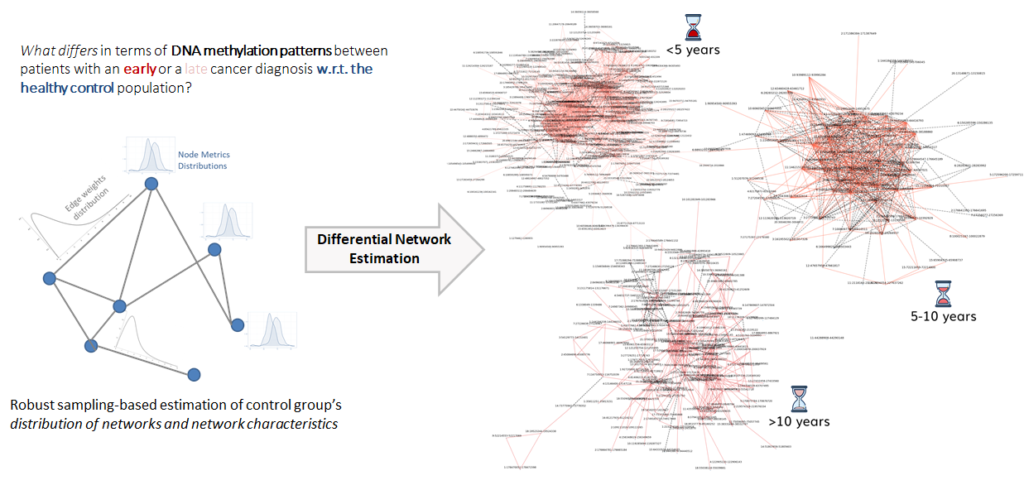
 Goals: : Identification of blood-based DNA methylation (DNAm) profiles predictive of cancer time to diagnosis (TTD), to improve the reliability/reproducibility of the results, as well as their biological meaningfulness. We will propose a shift from standard Epigenome-wide Association Studies (EWAS) that consider independent DNAm effects, and apply advanced Deep-Learning based models for survival analysis, coupled with Explanation Methods to model complex DNAm interrelationships and infer the global effect of DNAm sites all together on the phenotype.
Goals: : Identification of blood-based DNA methylation (DNAm) profiles predictive of cancer time to diagnosis (TTD), to improve the reliability/reproducibility of the results, as well as their biological meaningfulness. We will propose a shift from standard Epigenome-wide Association Studies (EWAS) that consider independent DNAm effects, and apply advanced Deep-Learning based models for survival analysis, coupled with Explanation Methods to model complex DNAm interrelationships and infer the global effect of DNAm sites all together on the phenotype.
Partners: Dr. Giovanni Fiorito (University of Sassari)
- EPIC2 – Multi-dimensional DNAm biomarkers creation via penalized mixed-effects multitask learning

Goals: DNA methylation (DNAm) is an epigenetic process that regulates gene expression, typically occurring in cytosines within CpG sites in the DNA sequence. In recent years, impressive epidemiological evidence has been established between DNAm epimutations and long-term exposure to lifestyle and environmental risk factors. To this extent, univariate multi-CpG DNAm biomarkers have been devised to predict patient-specific state of health indicators.
The project aims at creating a multi-dimensional DNAm biomarker of hypertension and hyperlipidemia from a multi-centric study. The learning mechanism is effectively constructed to accommodate custom penalty types coupled with random coefficients in a high-dimensional framework, achieving both flexibility and computational efficiency.
Partners: Dr. Giovanni Fiorito (University of Sassari)
- RAD Precise – Personalized radiotherapy: incorporating cellular response to irradiation in personalized treatment planning to minimize radiation toxicity (WP Partner of INT)
Goals: The aim of this collaborative translational project is to personalize radiotherapy (RT) treatment for cancer patients by incorporating information from improved predictive models for RT-induced toxicity based on data from multiple biomarkers of individual radiosensitivity into treatment planning systems (TPS). The goal is to tailor RT at the individual patient level by minimizing toxicity while maximizing tumour control.
The project exploits a prospective patient cohort established in the REQUITE project (www.requite.eu), with breast and prostate cancer patients and standardized collection of clinical, dosimetric and toxicity data, biobank and a minimum of two years follow-up. RADprecise take advantage of parametric models and machine learning to integrate the new biological data as well as already available genomics data to develop prediction models for RT-related adverse effects, which will be validated in an independent sample. Algorithms will be integrated into planning systems for biologically extended TPS. Health economics analyses will be conducted.
The project is interdisciplinary, including leading clinical scientists from academia and health research, Small & Medium Enterprises as well as patient advocates. Most collaborators have already cooperated on related topics, which provide the basis for RADprecise to take biological optimisation into the clinic.
Partners: Dr. Tiziana Rancati – PI (Istituto Nazionale dei Tumori), Dr. Alessandro Cicchetto, Prof. Paolo Zunino, Prof. Andrea Manzoni
- Polygenic Risk Scoring for Radiotherapy-induced Late Toxicity
 Goals: To identify the effect of single nucleotide polymorphism (SNP) interactions on the risk of toxicity following radiotherapy for prostate cancer and propose a new method for polygenic risk score incorporating SNP-SNP interactions (PRSi).
Goals: To identify the effect of single nucleotide polymorphism (SNP) interactions on the risk of toxicity following radiotherapy for prostate cancer and propose a new method for polygenic risk score incorporating SNP-SNP interactions (PRSi).
Partners: Dr. Tiziana Rancati – PI (Istituto Nazionale dei Tumori), Prof. Paolo Zunino, Prof. Andrea Manzoni
- Impact of Host Genome on COVID-19 clinical variability (partnership of University of Siena)

Goals: Our contribution to the project is the development of a Differential Network-based algorithm to characterize COVID-19 patients with severe outcomes and identify the genetic variants’ interaction patterns that differentiate this population from the rest of the patients. Moreover, the algorithm will allow us to further investigate these individuals by identifying the differential sets of variants significantly associated to the organ damages these severe cases suffered in acute phase, providing an additional stratification of variants and their roles in determining the phenotype.
Partners: Prof. Alessandra Renieri – PI, Dr. Chiara Fallerini (University of Siena)
People involved:
- Prof. Francesca Ieva
- Prof. Anna Paganoni
- Dr. Chiara Masci
- Dr. Andrea Cappozzo
- Dr. Lara Cavinato
- Dr. Michela Massi
- Dr. Nicola Rares-Franco

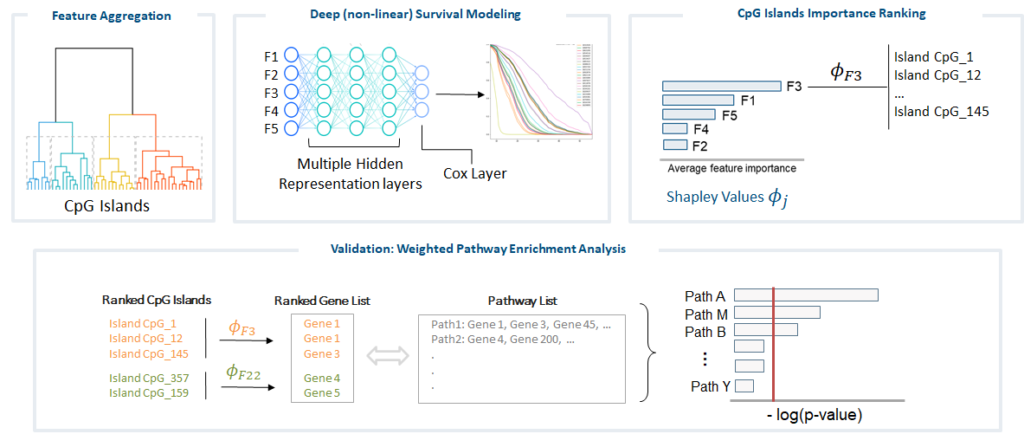 Goals: : Identification of blood-based DNA methylation (DNAm) profiles predictive of cancer time to diagnosis (TTD), to improve the reliability/reproducibility of the results, as well as their biological meaningfulness. We will propose a shift from standard Epigenome-wide Association Studies (EWAS) that consider independent DNAm effects, and apply advanced Deep-Learning based models for survival analysis, coupled with Explanation Methods to model complex DNAm interrelationships and infer the global effect of DNAm sites all together on the phenotype.
Goals: : Identification of blood-based DNA methylation (DNAm) profiles predictive of cancer time to diagnosis (TTD), to improve the reliability/reproducibility of the results, as well as their biological meaningfulness. We will propose a shift from standard Epigenome-wide Association Studies (EWAS) that consider independent DNAm effects, and apply advanced Deep-Learning based models for survival analysis, coupled with Explanation Methods to model complex DNAm interrelationships and infer the global effect of DNAm sites all together on the phenotype.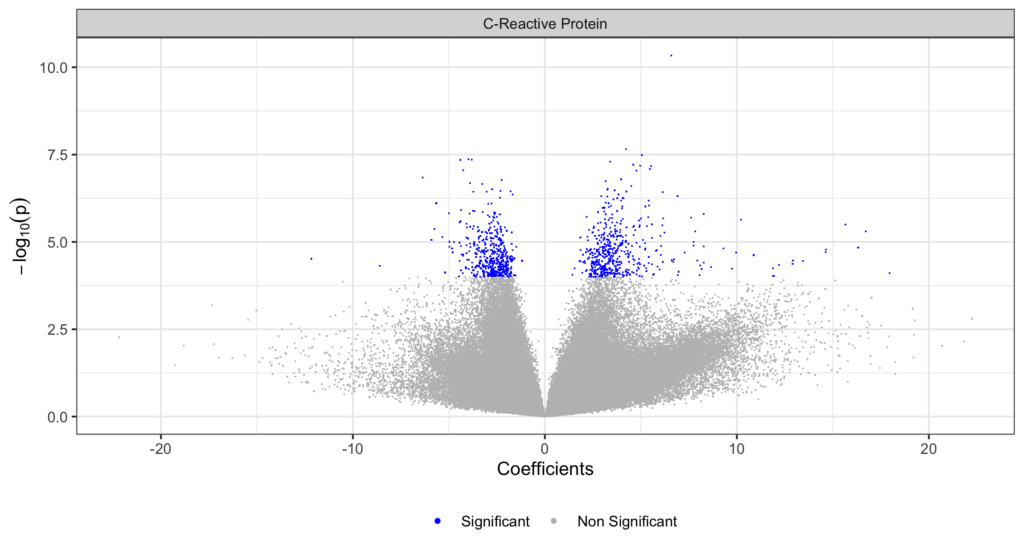
 Goals: To identify the effect of single nucleotide polymorphism (SNP) interactions on the risk of toxicity following radiotherapy for prostate cancer and propose a new method for polygenic risk score incorporating SNP-SNP interactions (PRSi).
Goals: To identify the effect of single nucleotide polymorphism (SNP) interactions on the risk of toxicity following radiotherapy for prostate cancer and propose a new method for polygenic risk score incorporating SNP-SNP interactions (PRSi).